![]()
|
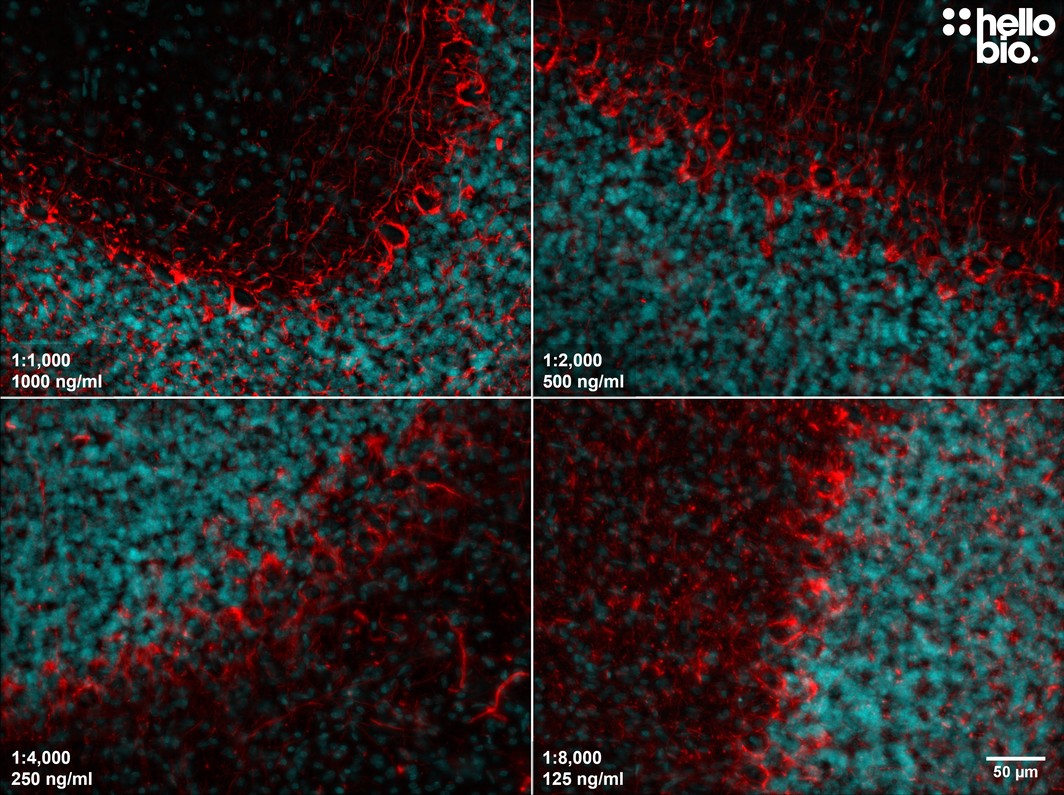
Figure 7. The effect of varying HB7266 concentration upon staining in rat cerebellum. HB7266 produced visible NFL staining at as low a concentration as 125ng/ml (1:8,000 dilution). Method: Brains were dissected from adult rats and fixed for 48hrs in 4% PFA before then incubated in 30% sucrose (in PBS) until the brains had sunk. A freezing microtome was used to cut 40µm transverse slices before sections were incubated in 1% NaBH4 for 30 minutes followed by 0.05M glycine for 30 minutes. Sections were blocked in 2% BSA, 3% goat serum for 2 hours before incubation overnight in HB7266 ranging in concentration from 0.125 to 1µg/ml (1:8,000 to 1:1000) at 4°C. This was followed by a two hour incubation with secondary antibody (polyclonal goat anti-rabbit DyLight 594 conjugated, Thermofisher 35561, 1:300 dilution). DAPI (HB0747) was used at 1µg/ml to visualise cell nuclei. For more detail please see our IHC(IF) protocol. Images were captured using a Leica DMi8 inverted epifluorescence microscope (20x objective) in a z-stack (1µm spacing) coupled to a Leica DFC365FX monochrome digital camera with DAPI LP and RHOD LP or Y5 filters. Exposure times were as follows:
1:1000 – DAPI 23.8ms (1x gain), RHOD 87.0ms (3.2x gain)
1:2000 – DAPI 17.3ms (1x gain), Y5 408.5ms (2.2x gain)
1:4000 – DAPI 17.5ms (1x gain), RHOD 101.3ms (2.1x gain)
1:8000 – DAPI 11.9ms (1x gain), RHOD 408.5ms (1.1x gain)
Images were processed in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682) using the subtract background (75px rolling ball radius) tool followed by Z-projection, stacking and montage creation. |
![]()
|
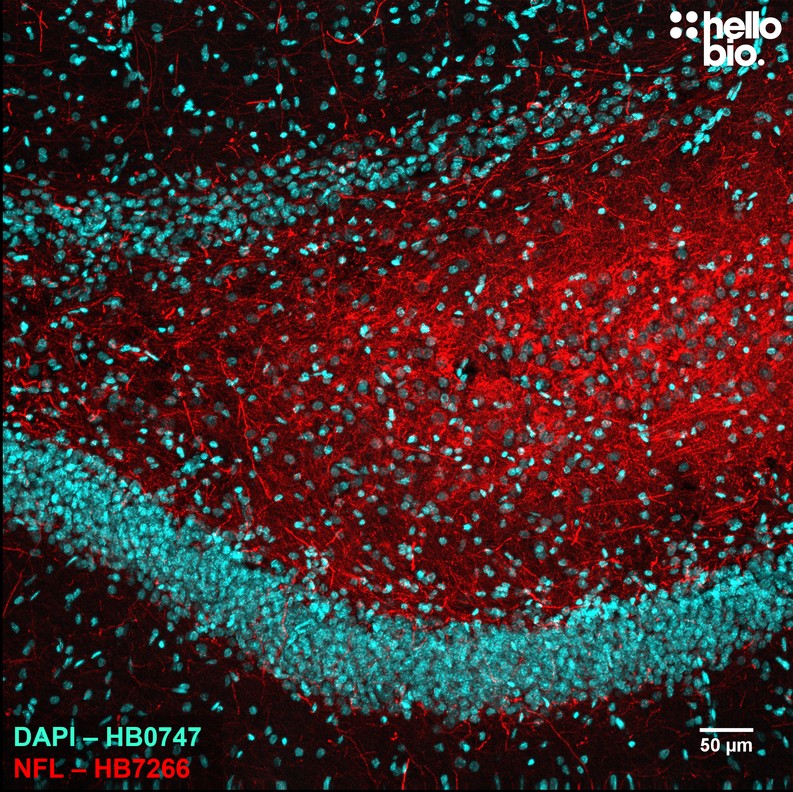
Figure 3. Neurofilament L expression in rat dorsal hippocampus visualised using HB7266. HB7266 visualised the mossy fibre projections from dentate granule cells to CA3 pyramidal neurones in the rat dentate gyrus. Method: Brains were dissected from adult rats and fixed for 48hrs in 4% PFA before then incubated in 30% sucrose (in PBS) until the brains had sunk. A freezing microtome was used to cut 40µm transverse slices before sections were incubated in 1% NaBH4 for 30 minutes followed by 0.05M glycine for 30 minutes. Sections were blocked in 2% BSA, 3% goat serum before incubation overnight in HB7266 (1:4000, 0.25µg/ml) at 4°C. This was followed by a two hour incubation with a polyclonal goat anti-rabbit DyLight 594 conjugated antibody (Thermofisher 35561, 1:300 dilution). DAPI (HB0747) was used at 1µg/ml to visualise cell nuclei. For more detail please see our IHC(IF) protocol . Images were captured using a Leica SPE confocal laser scanning microscope coupled to a Leica DMi8 inverted epifluorescence microscope. The image was captured using a 10x objective, 405nm (31.2% power, gain: 643) and 532nm laser lines (32.3% power, gain: 864) in a z-stack (3.25 µm spacing). Deconvolution was carried out using Huygens Essential (Scientific Volume Imagine) followed by the stack being flattened using a maximum Z projection in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682). |
![]()
|
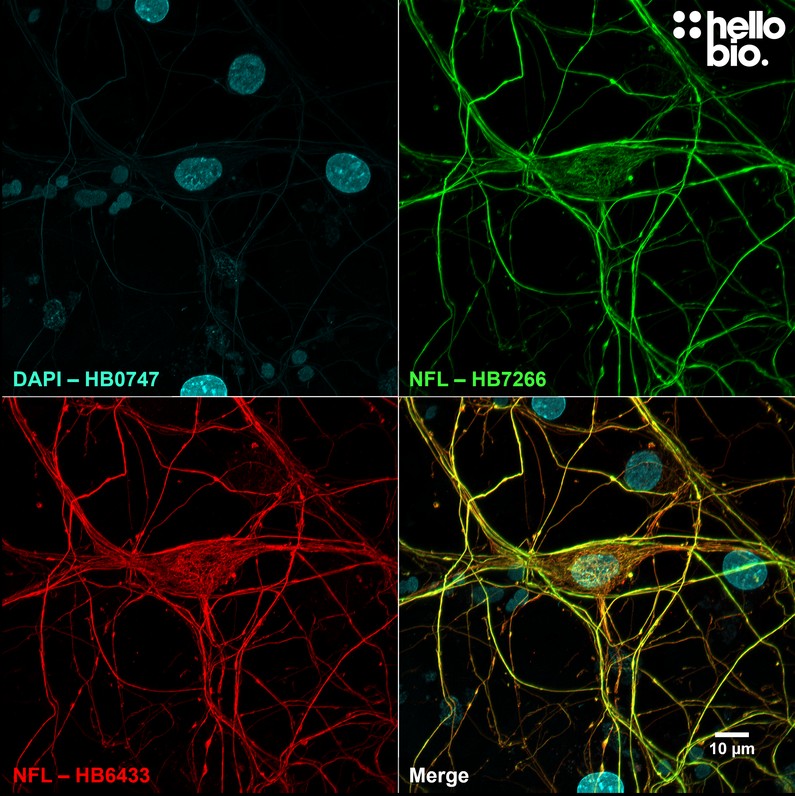
Figure 1. Independent antibody validation of HB7266 in cultured rat neurons. HB7266 (rabbit monoclonal antibody) and HB6433 (mouse monoclonal antibody) staining co-localises therefore shows strong evidence for the specificity of both antibodies. Method: neurones were cultured from PND2 rats following established protocols (Brewer and Torricelli, 2007. Nat Protoc 2, 1490–1498) and fixed with 4% PFA on DIV21. Cells were permeabilised with 0.1% Triton X-100 followed by blocking in 1% BSA, 300mM glycine. HB7266 (1:2000, 0.5µg/ml) and HB6433 (1:1000, 1µg/ml) were incubated overnight at 4°C followed by a one hour incubation with secondary antibodies (Polyclonal goat anti-mouse DyLight 594 conjugated, Thermofisher 35511, 1:300 dilution and polyclonal goat anti-rabbit DyLight 488 conjugated, Thermofisher 35552, 1:300 dilution). DAPI (HB0747) was used at 1µg/ml to visualise cell nuclei. For more detail please see our ICC protocol. Images were captured using a Leica SPE confocal laser scanning microscope coupled to a Leica DMi8 inverted epifluorescence microscope. The image was captured using a 63x objective, 405nm (28.9% power, gain: 624), 488nm (28.9% power, gain: 564) and 532nm laser lines (28.9% power, gain: 771) in a z-stack (0.35 µm spacing). Deconvolution was carried out using Huygens Essential version (Scientific Volume Imagine) followed by the stack being flattened using a maximum Z projection in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682). |
![]()
|
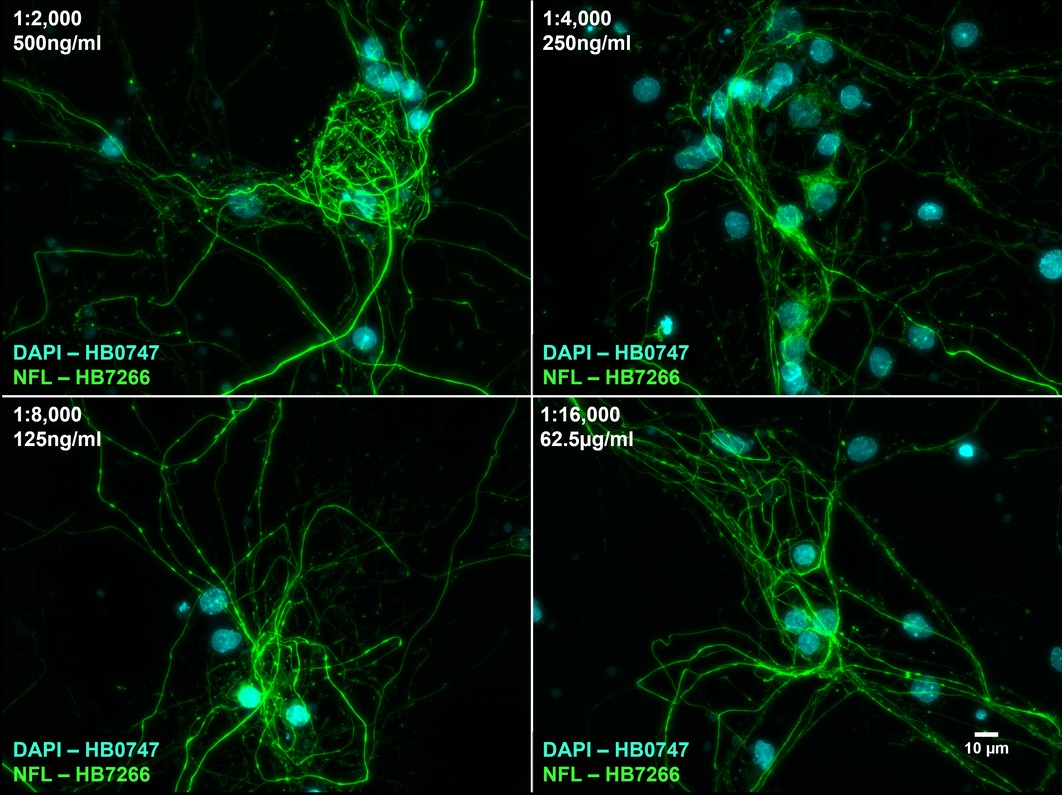
Figure 8. The effect of varying HB7266 concentration upon NFL staining in cultured rat neurones. HB7266 produces consistent staining with a high signal to noise ratio at concentrations as low at 62.5ng/ml (1:16,000 dilution). Method: neurones were cultured from PND2 rats following established protocols (Brewer and Torricelli, 2007. Nat Protoc 2, 1490–1498) and fixed with 4% PFA on DIV21. Cells were permeabilised with 0.1% Triton X-100 followed by blocking in 1% BSA, 300mM glycine. HB7266 was incubated overnight (4°C) at dilutions ranging from 1:2000 (500ng/ml) to 1:16,000 (125ng/ml). This was followed by a one hour incubation with secondary antibody (Polyclonal goat anti-rabbit DyLight 488 conjugated, Thermofisher 35561, 1:300 dilution). DAPI (HB0747) was used at 1µg/ml to visualise cell nuclei. For more detail please see our ICC protocol. Images were captured using a Leica DMi8 inverted epifluorescence microscope (40x objective) in a z-stack (1µm spacing) coupled to a Leica DFC365FX monochrome digital camera with DAPI LP and FITC LP filters. Exposure times were as follows:
1:2,000 – DAPI 125.3ms (1x gain), FITC 322.1ms (1x gain)
1:4,000 – DAPI 73.5ms (1x gain), FITC 322.1ms (1x gain)
1:8,000 – DAPI 73.5ms (1x gain), FITC 193.9ms (1x gain)
1:16,000 – DAPI 73.5ms (1x gain), FITC 286.3ms (1x gain)
Images were processed in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682) using the subtract background (50px rolling ball radius) tool followed by Z-projection, stacking and montage creation. |
![]()
|
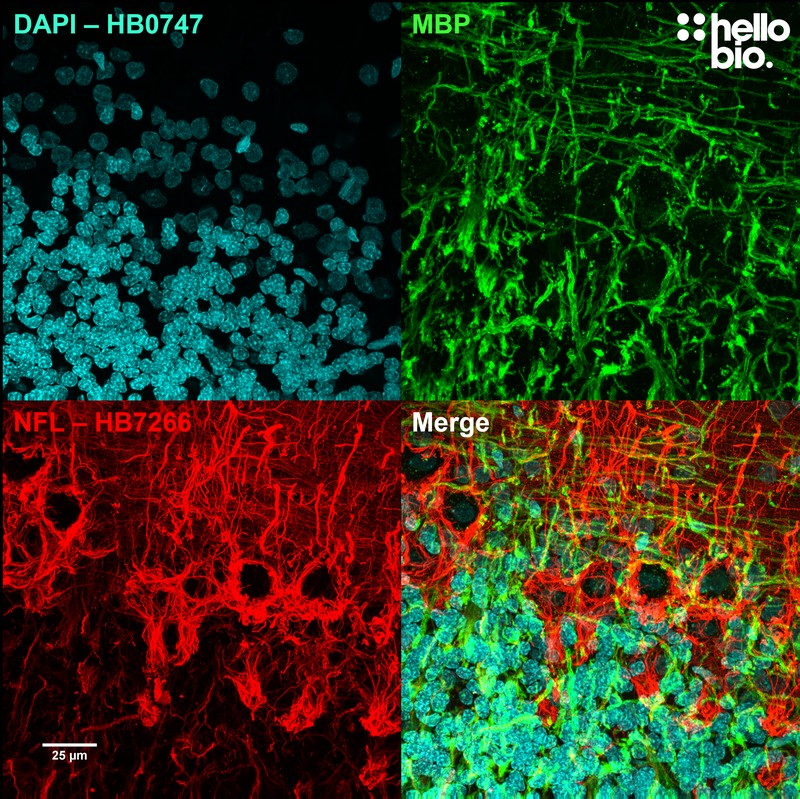
Figure 2. Neurofilament L and MBP staining in rat cerebellum visualised using HB7266. HB7266 visualised the dense network of neuronal projections in the cerebellum. Method: Brains were dissected from adult rats and fixed for 48hrs in 4% PFA before then incubated in 30% sucrose (in PBS) until the brains had sunk. A freezing microtome was used to cut 40µm transverse slices before sections were incubated in 1% NaBH4 for 30 minutes followed by 0.05M glycine for 30 minutes. Sections were blocked in 2% BSA, 3% goat serum before incubation overnight in HB7266 (1:2000, 0.5µg/ml) and an anti-MBP antibody (1:2000, 0.5µg/ml) at 4°C. This was followed by a two hour incubation with secondary antibodies (Polyclonal goat anti-mouse DyLight 488 conjugated, Thermofisher 35503, 1:300 dilution and polyclonal goat anti-rabbit DyLight 594 conjugated, Thermofisher 35561, 1:300 dilution). DAPI (HB0747) was used at 1µg/ml to visualise cell nuclei. For more detail please see our IHC(IF) protocol . Images were captured using a Leica SPE confocal laser scanning microscope coupled to a Leica DMi8 inverted epifluorescence microscope. The image was captured using a 40x objective, 405nm (31.2% power, gain: 578), 488nm (31.2% power, gain: 793) and 532nm laser lines (32.3% power, gain: 750) in a z-stack (0.33 µm spacing). Deconvolution was carried out using Huygens Essential (Scientific Volume Imagine) followed by the stack being flattened using a maximum Z projection in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682). |
![]()
|
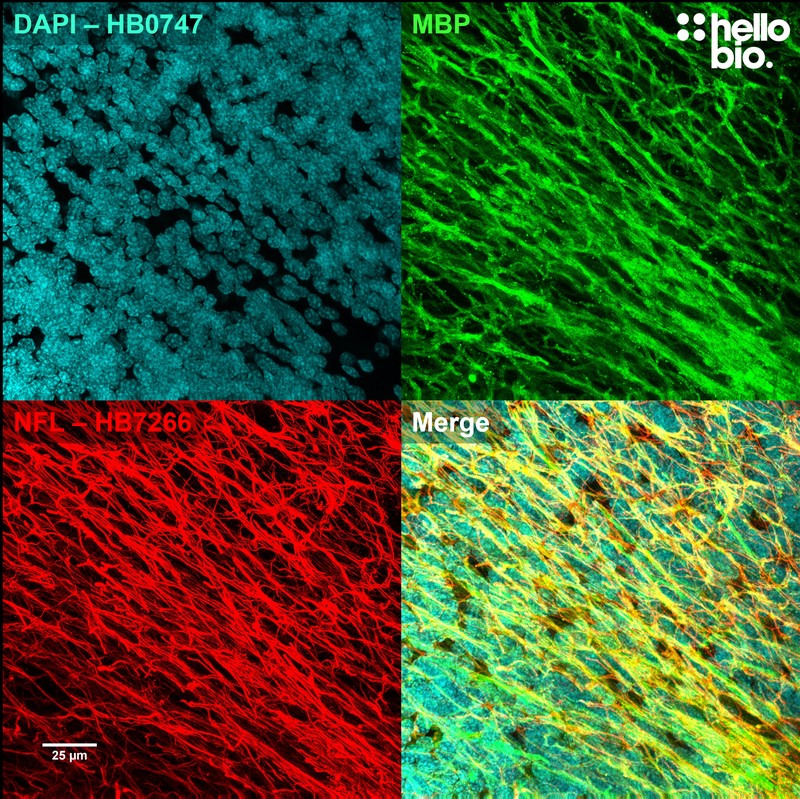
Figure 4. Neurofilament L and MBP staining in rat cerebellum visualised using HB7266. HB7266 visualised the dense network of fibres in the cerebellum. Method: Brains were dissected from adult rats and fixed for 48hrs in 4% PFA before then incubated in 30% sucrose (in PBS) until the brains had sunk. A freezing microtome was used to cut 40µm transverse slices before sections were incubated in 1% NaBH4 for 30 minutes followed by 0.05M glycine for 30 minutes. Sections were blocked in 2% BSA, 3% goat serum before incubation overnight in HB7266 (1:1000, 1µg/ml) and an anti-MBP antibody (1:1000, 1µg/ml) at 4°C. This was followed by a two hour incubation with secondary antibodies (Polyclonal goat anti-mouse DyLight 488 conjugated, Thermofisher 35503, 1:300 dilution and polyclonal goat anti-rabbit DyLight 594 conjugated, Thermofisher 35561, 1:300 dilution). DAPI (HB0747) was used at 1µg/ml to visualise cell nuclei. For more detail please see our IHC(IF) protocol . Images were captured using a Leica SPE confocal laser scanning microscope coupled to a Leica DMi8 inverted epifluorescence microscope. The image was captured using a 40x objective, 405nm (31.2% power, gain: 578), 488nm (31.2% power, gain: 793) and 532nm laser lines (32.3% power, gain: 750) in a z-stack (0.34 µm spacing). Deconvolution was carried out using Huygens Essential (Scientific Volume Imagine) followed by the stack being flattened using a maximum Z projection in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682). |
![]()
|
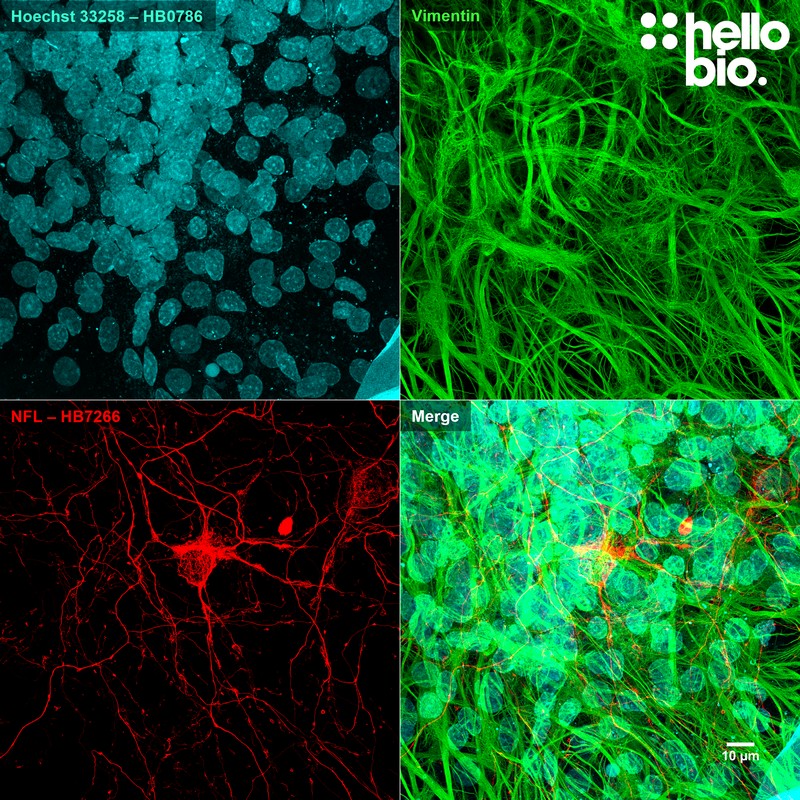
Figure 9. Neurofilament L (HB7266) and Vimentin staining in cultured rat neurones. Staining for vimentin and neurofilament L reveals the dense network of glial cells surrounding a neuron in culture. Method: neurones were cultured from PND2 rats following established protocols (Brewer and Torricelli, 2007. Nat Protoc 2, 1490–1498) and fixed with 4% PFA on DIV21. Cells were permeabilised with 0.1% Triton X-100 followed by blocking in 1% BSA, 300mM glycine. HB7266 (1:4000, 0.25µg/ml) and a mouse monoclonal anti-vimentin antibody (1:1000, 1µg/ml) were incubated overnight at 4°C followed by a one hour incubation with secondary antibodies (Polyclonal goat anti-mouse DyLight 488 conjugated, Thermofisher 35503, 1:300 dilution and polyclonal goat anti-rabbit DyLight 594 conjugated, Thermofisher 35561, 1:300 dilution). Hoechst 33258 (HB0786) was used at 10µg/ml for 10 mins to visualise nuclei. For more detail please see our ICC protocol. Images were captured using a Leica SP8 AOBS confocal laser scanning microscope attached to a Leica DMi8 inverted epifluorescence microscope. The image was captured using Lightning adaptive deconvolution using a 63x objective and 405nm (20.0% power, PMT: 907.4V gain), 496nm (0.5% power, HyD: 15.5% gain) and 561nm (0.5% power, HyD: 24.7% gain) lasers. Images were captured as a stack (0.37µm z-spacing) being flattened using a maximum Z projection in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682). |
![]()
|
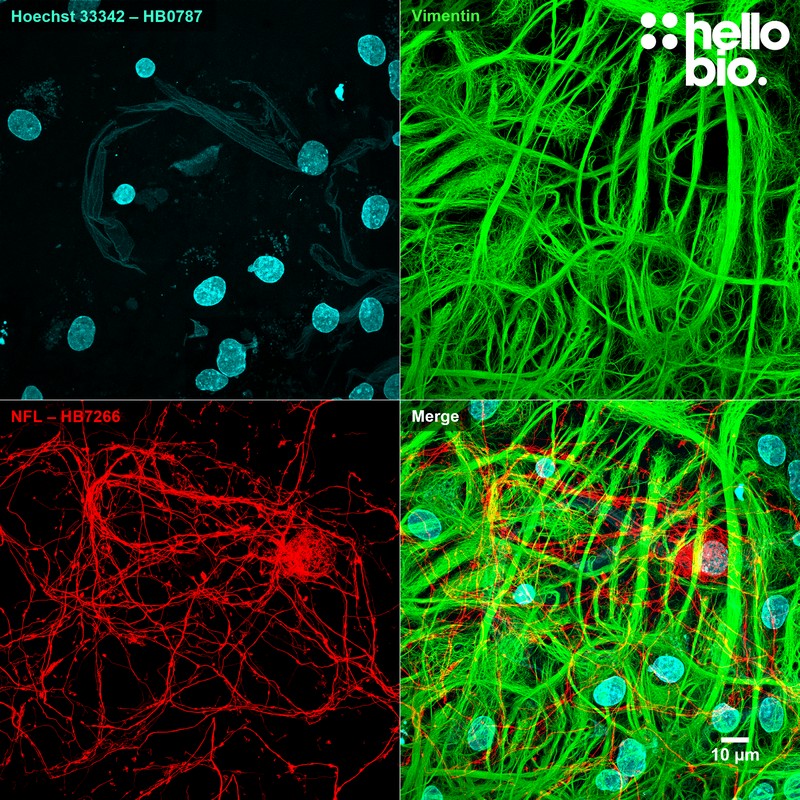
Figure 10. Neurofilament L (HB7266) and Vimentin staining in cultured rat neurones. Staining for vimentin and neurofilament L reveals the dense network of glial cells surrounding a neuron in culture. Method: neurones were cultured from PND2 rats following established protocols (Brewer and Torricelli, 2007. Nat Protoc 2, 1490–1498) and fixed with 4% PFA on DIV21. Cells were permeabilised with 0.1% Triton X-100 followed by blocking in 1% BSA, 300mM glycine. HB7266 (1:4000, 0.25µg/ml) and a mouse monoclonal anti-vimentin antibody (1:1000, 1µg/ml) were incubated overnight at 4°C followed by a one hour incubation with secondary antibodies (Polyclonal goat anti-mouse DyLight 488 conjugated, Thermofisher 35503, 1:300 dilution and polyclonal goat anti-rabbit DyLight 594 conjugated, Thermofisher 35561, 1:300 dilution). Hoechst 33342 (HB0787) was used at 1µg/ml for 10 mins to visualise nuclei. For more detail please see our ICC protocol. Images were captured using a Leica SP8 AOBS confocal laser scanning microscope attached to a Leica DMi8 inverted epifluorescence microscope. The image was captured using Lightning adaptive deconvolution using a 63x objective and 405nm (20.0% power, PMT: 893.7V gain), 496nm (0.5% power, HyD: 10% gain) and 561nm (0.5% power, HyD: 14.3% gain) lasers. Images were captured as a stack (0.25µm z-spacing) being flattened using a maximum Z projection in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682). |
![]()
|
Figure 5. Neurofilament L expression in various tissue lysates and preparations. HB7266 revealed a single band of around 72kDa present only in brain cytosol fractions. Method: mouse brain and rat brain membrane (P2) and cytosol fractions were prepared following previous work (Molnar et al., 1993. Neuroscience 53:307-326) from freshly collected adult brains. Other tissue lysates were prepared following established protocols from freshly dissected tissue (see our guide on WB sample preparation). Samples were loaded (20µg / lane) onto a 12% acrylamide gel alongside a protein ladder (NEB colour prestained broad range, P7719S) before being run at 60V for 54 minutes followed by 130V for 106 minutes. Wet transfer to a PVDF membrane was completed in 90 minutes using 400mA. The membrane was blocked for 2hrs in 5% non-fat dry milk before being incubated overnight at 4°C in HB7266 at a 1:1,000,000 dilution (1ng/ml). Following washing the membrane was incubated in secondary antibody (1:10,000 dilution, Polyclonal goat anti-rabbit HRP conjugated, Sigma Aldrich A6154) for 2hrs. For more detail please see our Western blotting protocol. Detection was accomplished using Clarity Western ECL substrate (BioRad, 1705061) and a Licor Odyssey Fc imaging system (ECL channel: 10 min exposure, 700nm channel: 30 sec exposure). Following imaging the membrane was stripped with two changes of stripping buffer (15g/l glycine, 1g/l SDS, 1% Tween 20, pH2.2) before being washed, blocked for 2 hours in 5% non-fat dry milk and incubated in HB9177 (1:4,000 dilution, 0.25µg/ml) overnight at 4°C. Following washing the membrane was incubated in secondary antibody (1:10,000 dilution, Polyclonal goat anti-mouse HRP conjugated, Sigma Aldrich A3682) for 2hrs and visualised again using Clarity Western ECL substrate (BioRad, 1705061) and a Licor Odyssey Fc imaging system (ECL channel: 10 min exposure, 700nm channel: 30 sec exposure). |
![]()
|
Figure 6. Concentration response of HB7266 staining in a rat brain cytosol preparation. HB7266 shows extremely high affinity for neurofilament L with bands being visible at concentrations as low as 100pg/ml. At higher concentrations degradation bands of NFL are visible (>1.5ng/ml) while at excessively high concentrations there is a slight amount of cross-reactivity with NFH and NFM (only ≥25ng/ml) which disappears as the concentration of HB7266 decreases. Method: cytosol fractions were prepared from fresh rat brains following established protocols (Molnar et al., 1993. Neuroscience 53:307-326). Rat cytosol samples were loaded (20µg / lane) onto a 12% acrylamide gel alongside a protein ladder (NEB colour prestained broad range, P7719S) before being run at 60V for 30 minutes followed by 120V for 100 minutes. Wet transfer to a PVDF membrane was completed in 90 minutes using 400mA. Following transfer the membrane was cut into strips using Ponceau dye to visualise and cut individual lanes. Strips were blocked for 3hrs in 5% non-fat dry milk before being incubated overnight at 4°C in HB7266. Each strip was incubated separately with a separate HB7266 concentration with this ranging from 100ng/ml (1:10,000 dilution) to 12pg/ml (1:81,920,000 dilution). Following washing the membrane was incubated in secondary antibody (1:10,000 dilution, Polyclonal goat anti-rabbit HRP conjugated, Sigma Aldrich A6154) for 2hrs. For more detail please see our Western blotting protocol. Detection was accomplished using Clarity Western ECL substrate (BioRad, 1705061) and a Licor Odyssey Fc imaging system (ECL channel: 10 min exposure, 700nm channel: 30 sec exposure). Band intensity was calculated using Image Studio version 5.2.5 (LiCor) and a graph was constructed in GraphPad Prism 9 using a 3-parameter Hill equation curve fit. |