![]()
|
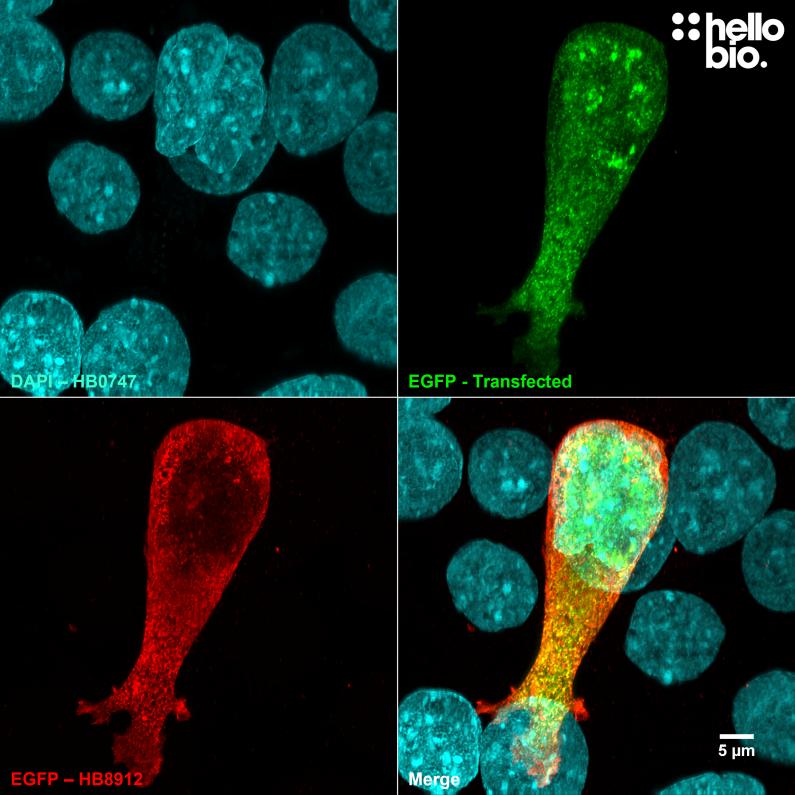
Figure 1. pEGFP-C2 transfected HEK293 cells showing co-localised staining of EGFP and HB8912. Signals derived from EGFP and HB8912 completely overlap showing specificity. Method: HEK293 cells were cultured and transfected following established protocols (Lee et al., 2019. PLoS ONE, 14(5):e0213116) with a pEGFP-C2 plasmid. After allowing cells time to express fixation was carried out using 4% PFA. Cells were permeabilised with 0.1% Triton X-100 followed by blocking in 1% BSA, 300mM glycine. HB8912 was incubated overnight (4°C) at a 1:1000 dilution (1µg/ml) followed by a one hour incubation with secondary antibody (Polyclonal goat anti-rabbit DyLight 594 conjugated, Thermofisher 35561, 1:300 dilution). DAPI (HB0747) was used at 1µg/ml to visualise cell nuclei. For more detail please see our ICC protocol. Images were captured using a Leica SPE confocal laser scanning microscope coupled to a Leica DMi8 inverted epifluorescence microscope. The image was captured using a 63x objective, 405nm (29.0% power), 488nm (37.9% power) and 532nm (36.8% power) laser lines in a z-stack (0.28 µm spacing). Deconvolution was carried out using Huygens Essential (Scientific Volume Imagine) followed by the stack being flattened using a maximum Z projection in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682). |
![]()
|
Figure 2. Specific HB8912 staining only in pEGFP-C2 transfected HEK293 cells. HB8912 revealed a primary 31.5kDa band only present in pEGFP-C2 transfected cells without any cross-reactivity with mCherry or native proteins. Method: HEK293 cells were cultured and transfected following established protocols (Lee et al., 2019. PLoS ONE, 14(5):e0213116) with either pEGFP-C2 or pmCherry-C3 plasmids. After allowing 3 days for expression lysates were prepared (see our guide on WB sample preparation) and loaded (equal loading) onto a 15% acrylamide gel alongside a protein ladder (New England Biolabs, P7719S) before being run at 60V for 30 minutes followed by 130V for 120 minutes. Wet transfer to a PVDF membrane was completed in 90 minutes using 400mA. The membrane was blocked for 2hrs in 5% non-fat dry milk before being incubated overnight at 4°C in HB8912 (Polyclonal rabbit anti-GFP) at a 1:10,000 dilution (0.1µg/ml). Following washing the membrane was incubated in secondary antibody (1:10,000 dilution, Polyclonal goat anti-rabbit HRP conjugated, Sigma Aldrich A6154) for 2hrs. For more detail please see our Western blotting protocol. Detection was accomplished using Clarity Western ECL substrate (BioRad, 1705061) and a Licor Odyssey Fc imaging system (ECL channel: 10 min exposure, 700nm channel: 30 sec exposure). Following imaging the membrane was stripped with two changes of stripping buffer (15g/l glycine, 1g/l SDS, 1% Tween 20, pH2.2) before being washed, blocked for 2 hours in 5% non-fat dry milk and incubated in HB9177 (1:4,000 dilution, 0.25µg/ml) overnight at 4°C. Following washing the membrane was incubated in secondary antibody (1:10,000 dilution, Polyclonal goat anti-mouse HRP conjugated, Sigma Aldrich A3682) for 2hrs and visualised again using Clarity Western ECL substrate (BioRad, 1705061) and a Licor Odyssey Fc imaging system (ECL channel: 10 min exposure, 700nm channel: 30 sec exposure). |
![]()
|
Figure 3. Concentration response of HB8912 staining in pEGFP-C2 transfected HEK293 cells. HB8912 shows consistent results at dilutions as low as 1:64,000 (15.6 ng/ml). Method: HEK293 cells were cultured and transfected following established protocols (Lee et al., 2019. PLoS ONE, 14(5):e0213116) with a pEGFP-C2 plasmid. After allowing 3 days for expression lysate was prepared (see our guide on WB sample preparation) and loaded (equal loading) onto a 15% acrylamide gel alongside a protein ladder (New England Biolabs, P7719S) before being run at 60V for 30 minutes followed by 130V for 120 minutes. Wet transfer to a PVDF membrane was completed in 90 minutes using 400mA. Following transfer the membrane was cut into strips using Ponceau dye to visualise and cut individual lanes. Strips were blocked for 2hrs in 5% non-fat dry milk before being incubated overnight at 4°C in HB8912. Each strip was incubated separately with a separate HB8912 concentration with this ranging from 2µg/ml (1:500 dilution) to 7.8ng/ml (1:128,000 dilution). Following washing the membrane was incubated in secondary antibody (1:10,000 dilution, Polyclonal goat anti-rabbit HRP conjugated, Sigma Aldrich A6154) for 2hrs. For more detail please see our Western blotting protocol. Detection was accomplished using Clarity Western ECL substrate (BioRad, 1705061) and a Licor Odyssey Fc imaging system (ECL channel: 10 min exposure, 700nm channel: 30 sec exposure). Band intensity was calculated using Image Studio version 5.2.5 (LiCor) and a graph was constructed in GraphPad Prism 9 using a 3-parameter Hill equation curve fit. |
![]()
|
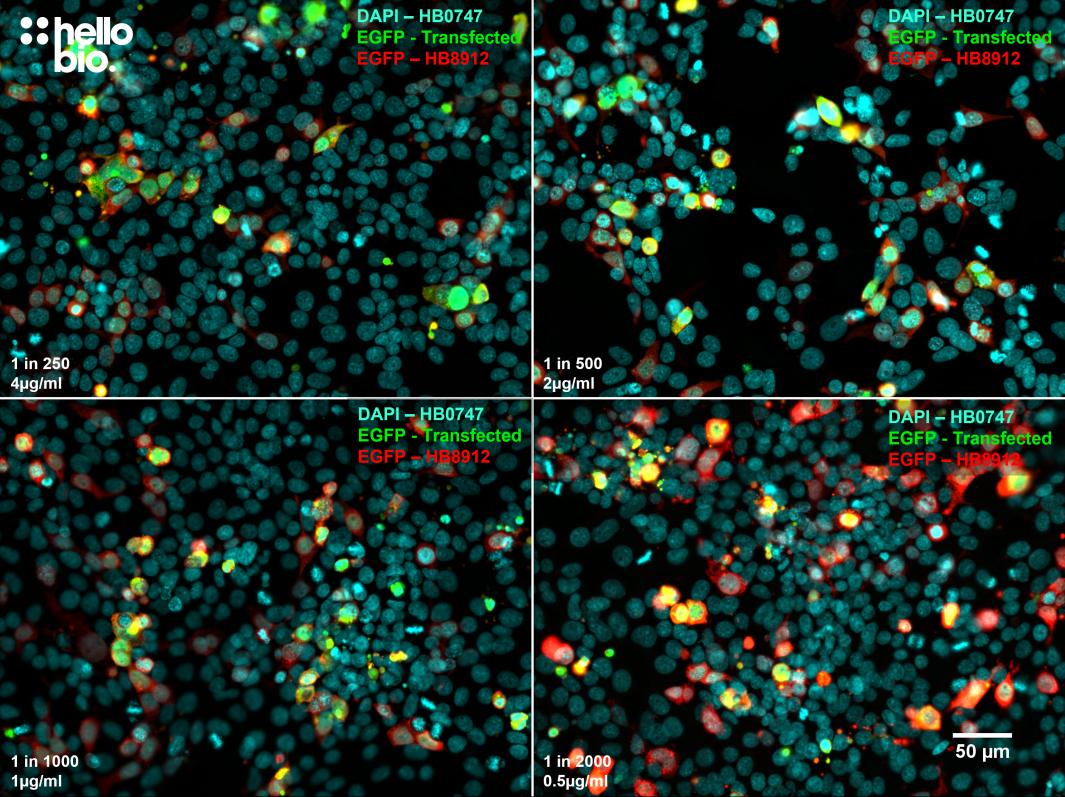
Figure 4. The effect of varying HB8912 concentration upon staining in pEGFP-C2 transfected HEK293 cells. HB8912 produced excellent amplification of native EGFP signal at concentrations as low as 0.5µg/ml (1:2000 dilution). Method: HEK293 cells were cultured and transfected following established protocols (Lee et al., 2019. PLoS ONE, 14(5):e0213116) with a pEGFP-C2 plasmid. After allowing cells time to express fixation was carried out using 4% PFA. Cells were permeabilised with 0.1% Triton X-100 followed by blocking in 1% BSA, 300mM glycine. HB8912 was incubated overnight (4°C) at concentrations ranging from 0.5 to 4µg/ml (1:2000 to 1:250) followed by a one hour incubation with secondary antibody (Polyclonal goat anti-rabbit DyLight 594 conjugated, Thermofisher 35561, 1:300 dilution). DAPI (HB0747) was used at 1µg/ml to visualise cell nuclei. For more detail please see our ICC protocol. Images were captured using a Leica DMi8 inverted epifluorescence microscope (20x objective) coupled to a Leica DFC365FX monochrome digital camera with DAPI LP, FITC LP and RHOD_LP filters. Exposure times were as follows:
1:250 DAPI LP: 1x gain, 49.8ms exposure; FITC LP: 2.5x gain, 142.3ms exposure; RHOD LP: 2x gain, 108.8ms exposure
1:500 DAPI LP: 1x gain, 49.8ms exposure; FITC LP: 1.2x gain, 117.2ms exposure; RHOD LP: 1.2x gain, 108.8ms exposure
1:1000 DAPI LP: 1x gain, 49.8ms exposure; FITC LP: 1.4x gain, 101.3ms exposure; RHOD LP: 1.2x gain, 246.0ms exposure
1:2000 DAPI LP: 1x gain, 49.8ms exposure; FITC LP: 1.4x gain, 86.6ms exposure; RHOD LP: 1.7x gain, 246.0ms exposure.
Images were processed in ImageJ (Schindelin et al., 2012. Nat Methods, 9(7), 676–682) using the subtract background (50px rolling ball radius) tool followed by stacking and montage creation. |
![]()
|
Figure 5. EGFP tagged Cannabinoid receptor 1 c-terminal domain (EGFP-ctCB1R) detected in virally transfected HEK293 cells using HB8912. HB8912 succesfully detected the shift in molecular weights when HEK293 cells were either transfected with a EGFP-ctCB1R construct or EGFP alone. Method: Sindbis virus was used to over-express EGFP-ctCB1R alongside EGFP control in HEK293 cells following the cited methodology (Fletcher Jones, 2020. PhD Thesis, University of Bristol). After allowing time for expression, lysates were prepared (see our guide on WB sample preparation) and loaded (equal loading) alongside a protein ladder (New England Biolabs, P7719S) onto a 10% acrylamide gel. The cytosol fraction from rat brains was also loaded as a non EGFP expressing control and was prepared following established methodology (Molnar et al., 1993. Neuroscience 53:307-326). Gels were run at 60V for 30 minutes followed by 120V for 100 minutes. Wet transfer to a PVDF membrane was completed in 90 minutes using 400mA. The membrane was blocked for 2hrs in 5% non-fat dry milk before being incubated overnight at 4°C in HB8912 (Polyclonal rabbit anti-GFP) at a 1:1,000 dilution (1µg/ml). Following washing the membrane was incubated in secondary antibody (1:10,000 dilution, Polyclonal goat anti-rabbit HRP conjugated, Sigma Aldrich A6154) for 2hrs. For more detail please see our Western blotting protocol. Detection was accomplished using Clarity Western ECL substrate (BioRad, 1705061) and a Licor Odyssey Fc imaging system (ECL channel: 10 min exposure, 700nm channel: 30 sec exposure). |